Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Schyman, P.*; Danielsson, J.*; Pinak, M.; Laaksonen, A.*
Journal of Physical Chemistry A, 109(8), p.1713 - 1719, 2005/02
Times Cited Count:39 Percentile:77.29(Chemistry, Physical)We have examined the role of the catalytic lysine (Lys 249) in breaking the glycosidic bond of 8-oxoguanine in the enzyme human 8-oxoguanine DNA glycosylase. It has been assumed that this lysine acts as a nucleophile in a S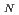 2 type of reaction after being activated through a donation of a proton to a strictly conserved aspartate. We use hybrid density functional theory to characterize both associative and dissociative pathways. We find that the smallest energetical barrier involves a S
2 type of reaction after being activated through a donation of a proton to a strictly conserved aspartate. We use hybrid density functional theory to characterize both associative and dissociative pathways. We find that the smallest energetical barrier involves a S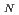 1 type of mechanism where the lysine electrostatically stabilizes the dissociating base and then donates a proton with a very small barrier and then finally attacks the sugar ring to create the covalently bounded protein-DNA intermediate complex. Reported findings give further support to the assumption that a dissociative mechanism may be the preferred mode of action for this type of enzymes.
1 type of mechanism where the lysine electrostatically stabilizes the dissociating base and then donates a proton with a very small barrier and then finally attacks the sugar ring to create the covalently bounded protein-DNA intermediate complex. Reported findings give further support to the assumption that a dissociative mechanism may be the preferred mode of action for this type of enzymes.
Pinak, M.
JAERI-Research 2002-016, 31 Pages, 2002/09
The molecular dynamics (MD) simulations of DNA mutagenic oxidative lesion 8-oxoG, single and complexed with the repair enzyme hOGG1, were performed. In the case of simulation of single DNA molecule the broken hydrogen bonds resulting in locally collapsed B-DNA structure were observed at the lesion site. In addition the adenine on the complementary strand was flipped-out of the DNA double helix. In the case of simulation of DNA and repair enzyme hOGG1, the DNA-enzyme complex was formed after 500 picoseconds of MD that lasted stable until the simulation was terminated at 1 ns. The N-terminus of arginine 313 was located close to the phosphodiester bond of nucleotide with 8-oxoG enabling chemical reactions between amino acid and lesion. Phosphodiester bond at C5'of 8-oxoG was at the position close to the N-terminus of arginine 313. The water mediated hydrogen bonds network was formed in each contact area between DNA and enzyme further enhancing the stability of complex.